Gitlab CI for Automated Environment Preservation
Overview
Teaching: 20 min
Exercises: 25 minQuestions
How can gitlab CI and docker work together to automatically preserve my analysis environment?
What do I need to add to my gitlab repo(s) to enable this automated environment preservation?
Objectives
Learn how to write a Dockerfile to containerize your analysis code and environment.
Understand what needs to be added to your
.gitlab-ci.ymlfile to keep the containerized environment continuously up to date for your repo.
Introduction
In this section, we learn how to combine the forces of docker and gitlab CI to automatically keep your analysis environment up-to-date. This is accomplished by adding an extra stage to the CI pipeline for each analysis repo, which builds a container image that includes all aspects of the environment needed to run the code.
Writing your Gitlab Dockerfile
The goal of automated environment preservation is to create a docker image that you can immediately start executing your analysis code inside upon startup. Let’s review the needed components for this.
- Set up the OS, system libraries, and other dependencies that your code depends on,
- Add your analysis code to the container, and
- Build the code so that it can just be executed trivially inside the container.
As we’ve seen, all these components can be encoded in a Dockerfile. So the first step to set up automated image building is to add a Dockerfile to the repo specifying these components.
The
rootproject/root-condadocker imageIn this tutorial, we build our analysis environments on top of the
rootproject/root-condabase image (link to project area on docker hub). This image comes with root 6.18 and python 3.7 pre-installed. It also comes with XrootD for downloading files from eos. Therootproject/root-condais itself built with a Dockerfile, which uses conda to install root and python on top of another base image (continuumio/miniconda3).
Exercise (15 min)
Working from your bash shell, cd into the top level of the repo you use for skimming. Create an empty file named
Dockerfile.touch DockerfileNow open the Dockerfile with a text editor and, starting with the following skeleton, fill in the FIXMEs to make a Dockerfile that fully specifies your analysis environment in this repo.
# Start from the rootproject/root-conda:6.18.04 base image [FIXME] # Put the current repo (the one in which this Dockerfile resides) in the /analysis/skim directory # Note that this directory is created on the fly and does not need to reside in the repo already [FIXME] # Make /analysis/skim the default working directory (again, it will create the directory if it doesn't already exist) [FIXME] # Compile an executable named 'skim' from the skim.cxx source file RUN echo ">>> Compile skimming executable ..." && \ COMPILER=[FIXME] && \ FLAGS=[FIXME] && \ [FIXME]Solution
# Start from the rootproject/root-conda base image FROM rootproject/root-conda:6.18.04 # Put the current repo (the one in which this Dockerfile resides) in the /analysis/skim directory # Note that this directory is created on the fly and does not need to reside in the repo already COPY . /analysis/skim # Make /analysis/skim the default working directory (again, it will create the directory if it doesn't already exist) WORKDIR /analysis/skim # Compile an executable named 'skim' from the skim.cxx source file RUN echo ">>> Compile skimming executable ..." && \ COMPILER=$(root-config --cxx) && \ FLAGS=$(root-config --cflags --libs) && \ $COMPILER -g -std=c++11 -O3 -Wall -Wextra -Wpedantic -o skim skim.cxx $FLAGSOnce you’re happy with your Dockerfile, you can commit it to your repo and push it to github.
Hints
As you’re working, you can test whether the Dockerfile builds successfully using the
docker buildcommand. Eg.docker build -t payload_analysis .When your image builds successfully, you can
runit and poke around to make sure it’s set up exactly as you want, and that you can successfully run the executable you built:docker run -it --rm payload_analysis /bin/bash
Now, you can proceed with updating your .gitlab-ci.yml to actually build the container during the CI/CD pipeline and store it in the gitlab registry. You can later pull it from the gitlab registry just as you would any other container, but in this case using your CERN credentials.
Add the following lines at the end of the .gitlab-ci.yml file to build the image and save it to the docker registry.
build_image:
stage: build
variables:
TO: $CI_REGISTRY_IMAGE:$CI_COMMIT_REF_SLUG-$CI_COMMIT_SHORT_SHA
tags:
- docker-image-build
script:
- ignore
Once this is done, you can commit and push the updated .gitlab-ci.yml file to your gitlab repo and check to make sure the pipeline passed. If it passed, the repo image built by the pipeline should now be stored on the docker registry, and be accessible as follows:
docker login gitlab-registry.cern.ch
docker pull gitlab-registry.cern.ch/[repo owner's username]/[skimming repo name]:[branch name]-[shortened commit SHA]
You can also go to the container registry on the gitlab UI to see all the images you’ve built:
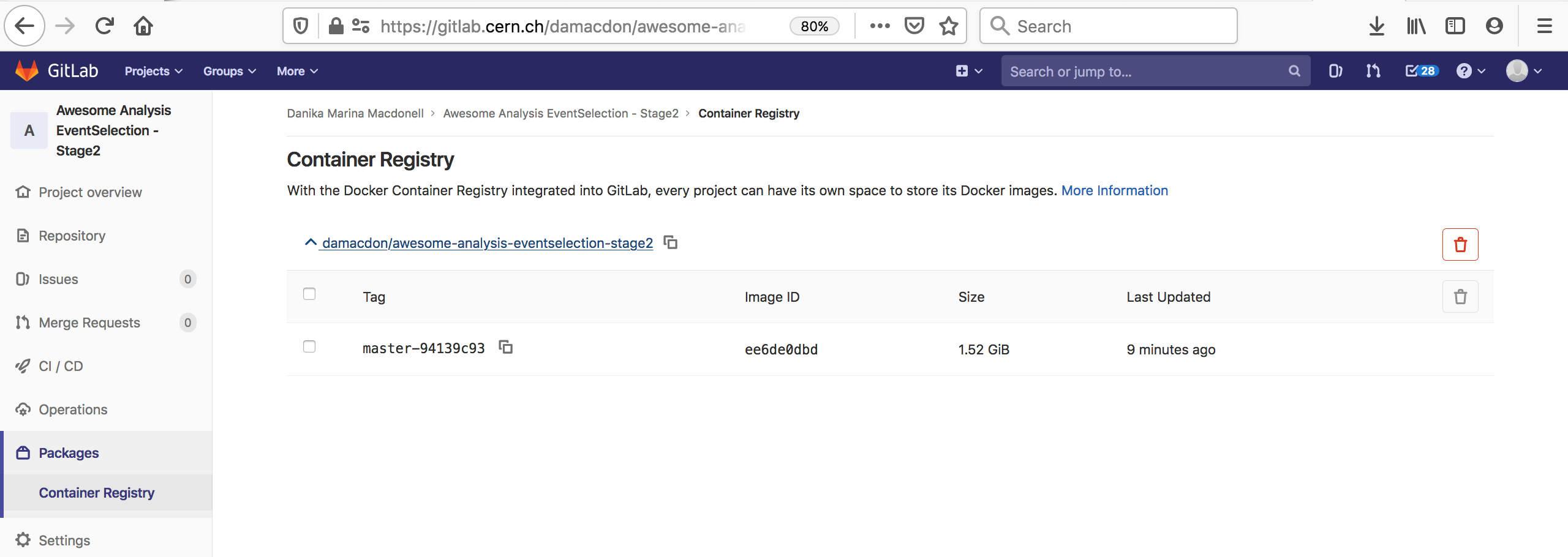
Notice that the script to run is just a dummy ‘ignore’ command. This is because using the docker-image-build tag, the jobs always land on special runners that are managed by CERN IT which run a custom script in the background. You can safely ignore the details.
Recommended Tag Structure
You’ll notice the environment variable
TOin the.gitlab-ci.ymlscript above. This controls the name of the Docker image that is produced in the CI step. Here, the image name will be<reponame>:<branch or tagname>-<short commit SHA>. The shortened 8-character commit SHA ensures that each image created from a different commit will be unique, and you can easily go back and find images from previous commits for debugging, etc.As you’ll see tomorrow, it’s recommended when using your images as part of a REANA workflow to make a unique image for each gitlab commit, because REANA will only attempt to update an image that it’s already pulled if it sees that there’s a new tag associated with the image.
If you feel it’s overkill for your specific use case to save a unique image for every commit, the
-$CI_COMMIT_SHORT_SHAcan be removed. Then the$CI_COMMIT_REF_SLUGwill at least ensure that images built from different branches will not overwrite each other, and tagged commits will correspond to tagged images.
Exercise (10 mins)
Since we’re now taking care of building the skimming executable during image building, let’s make an updated version of skim.sh that excludes the step of building the
skimexecutable.The updated script should just directly run the pre-existing
skimexecutable on the input samples. You could call it eg.skim_prebuilt.sh. We’ll be using this updated script in an exercise later on in which we’ll be going through the full analysis in containers launched from the images we create with gitlab CI.Once you’re happy with the script, you can commit and push it to the repo.
Solution
#!/bin/bash INPUT_DIR=$1 OUTPUT_DIR=$2 # Sanitize input path, XRootD breaks if we double accidentally a slash if [ "${INPUT_DIR: -1}" = "/" ]; then INPUT_DIR=${INPUT_DIR::-1} fi # Skim samples while IFS=, read -r SAMPLE XSEC do echo ">>> Skim sample ${SAMPLE}" INPUT=${INPUT_DIR}/${SAMPLE}.root OUTPUT=${OUTPUT_DIR}/${SAMPLE}Skim.root LUMI=11467.0 # Integrated luminosity of the unscaled dataset SCALE=0.1 # Same fraction as used to down-size the analysis ./skim $INPUT $OUTPUT $XSEC $LUMI $SCALE done < skim.csv
Key Points
gitlab CI allows you to re-build a container that encapsulates the environment each time new commits are pushed to the analysis repo.
This functionality is enabled by adding a Dockerfile to your repo that specifies how to build the environment, and an image-building stage to the .gitlab-ci.yml file.