Introduction
Overview
Teaching: 10 min
Exercises: 1 minQuestions
What makes research data analyses reproducible?
Is preserving code, data, and containers enough?
Objectives
Understand principles behind computational reproducibility
Understand the concept of serial and parallel computational workflows
Computational reproducibility
A reproducibility quote
An article about computational science in a scientific publication is not the scholarship itself, it is merely advertising of the scholarship. The actual scholarship is the complete software development environment and the complete set of instructions which generated the figures.
– Jonathan B. Buckheit and David L. Donoho, “WaveLab and Reproducible Research”, source
Computational reproducibility has many definitions. Example: The Turing Way definition of computational reproducibility: source
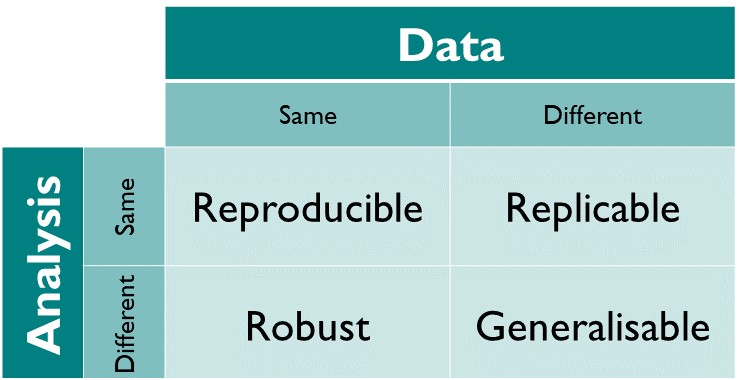
In other words: same data + same analysis = reproducible results
What about real life?
Example: Nature volume 533 issue 7604 (2016) surveying 1500 scientists. source
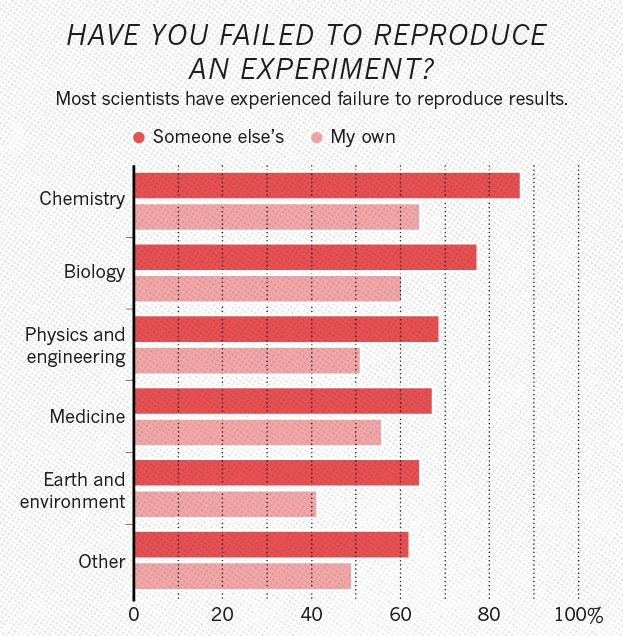
Half of researchers cannot reproduce their own results.
Slow uptake of best practices
Many “best practices” guidelines published. For example, “Ten Simple Rules for Reproducible Computational Research” by Geir Kjetil Sandve, Anton Nekrutenko, James Taylor, Eivind Hovig (2013) DOI:10.1371/journal.pcbi.1003285:
- For every result, keep track of how it was produced
- Avoid manual data manipulation steps
- Archive the exact versions of all external programs used
- Version control all custom scripts
- Record all intermediate results, when possible in standardized formats
- For analyses that include randomness, note underlying random seeds
- Always store raw data behind plots
- Generate hierarchical analysis output, allowing layers of increasing detail to be inspected
- Connect textual statements to underlying results
- Provide public access to scripts, runs, and results
Yet the uptake has been slow. Several reasons:
- sociological: publish-or-perish culture; missing incentives
- technological: easy-to-use tools
Top-down approaches (funding bodies asking for Data Management Plans) combined with bottom-up approaches (building tools integrating into daily research workflow) bringing the change.
A reproducibility quote
Your closest collaborator is you six monhts ago… and your younger self does not reply to emails.
Four questions
Four questions to aid robustness of analyses:
- Input data? Specify all input data and parameters.
- Analysis code? Specify all analysis code and libraries analysing the data.
- Compute environment? Specify all requisite liraries and operating system platform running the analysis.
- Runtime procedures? Specify all the computational steps taken to achieve the result.
Code and containerised environment was covered in previous two days; good!
Today we’ll cover the preservation of runtime procedures.
Exercise
Are containers enough to capture your runtime environment? What else might be necessary in your typical physics analysis scienarios?
Solution
Any external resources, such as database calls, must also be thought about. Will the external database that you use be there in two years?
Computational workflows
Use of interactive and graphical interfaces is not recommended, as one cannot reproduce user clicks easily.
Use of custom helper scripts (e.g. run.sh shell scripts) or custom orchestration scripts (e.g.
Python glue code) running the analysis is much better.
However, porting glue code to new usage scenarios may be tedious work that is better spent doing research.
Hence the birth of declarative workflow systems that express the computational steps more abstractly.
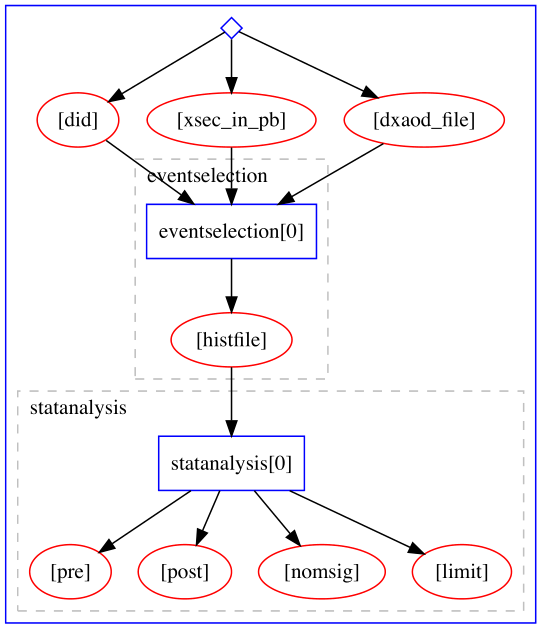
Example of a serial computational workflow typical for ATLAS RECAST analyses:
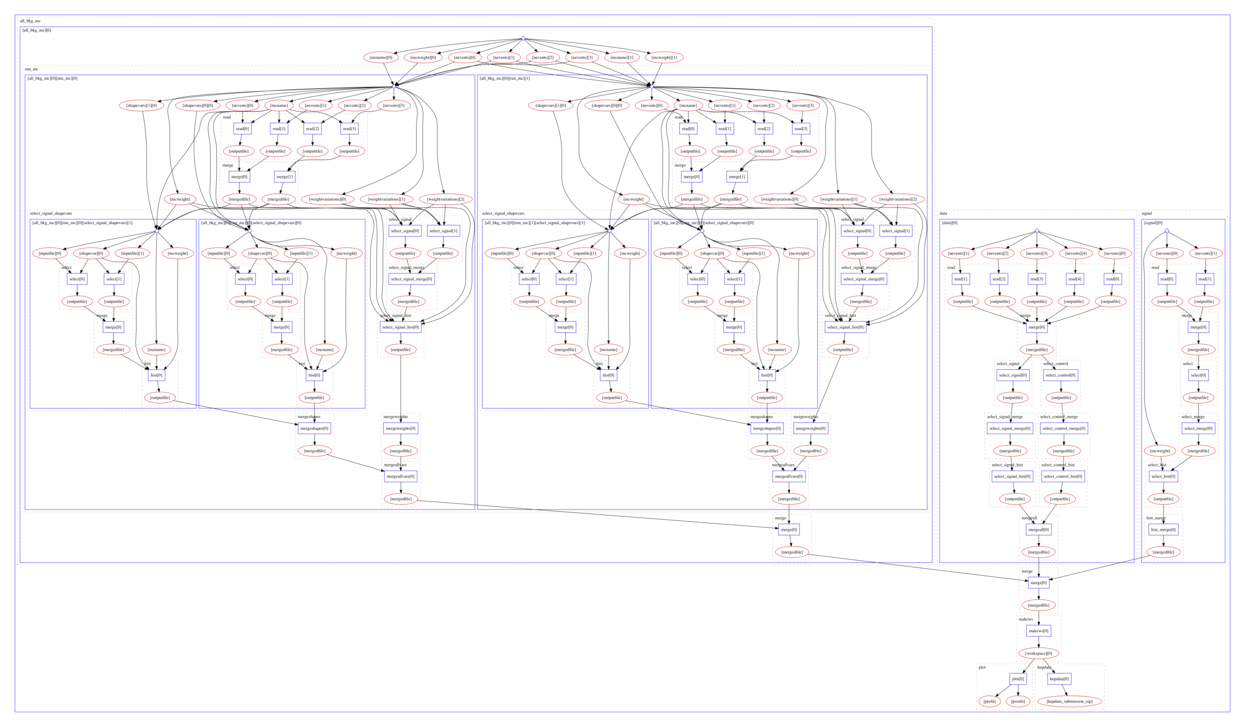
Example of a parallel computational workflow typical for Beyond Standard Model searches:
Many different computational data analysis workflow systems exist.
Different tools used in different communities: fit for use, fit for purpose, culture, preferences.
REANA
We shall use REANA reproducible analysis platform to explore computational workflows in this lesson. REANA is a pilot project and supports:
- multiple workflow systems (CWL, Serial, Yadage)
- multiple compute backends (Kubernetes, HTCondor, Slurm)
- multiple storage backends (Ceph, EOS)
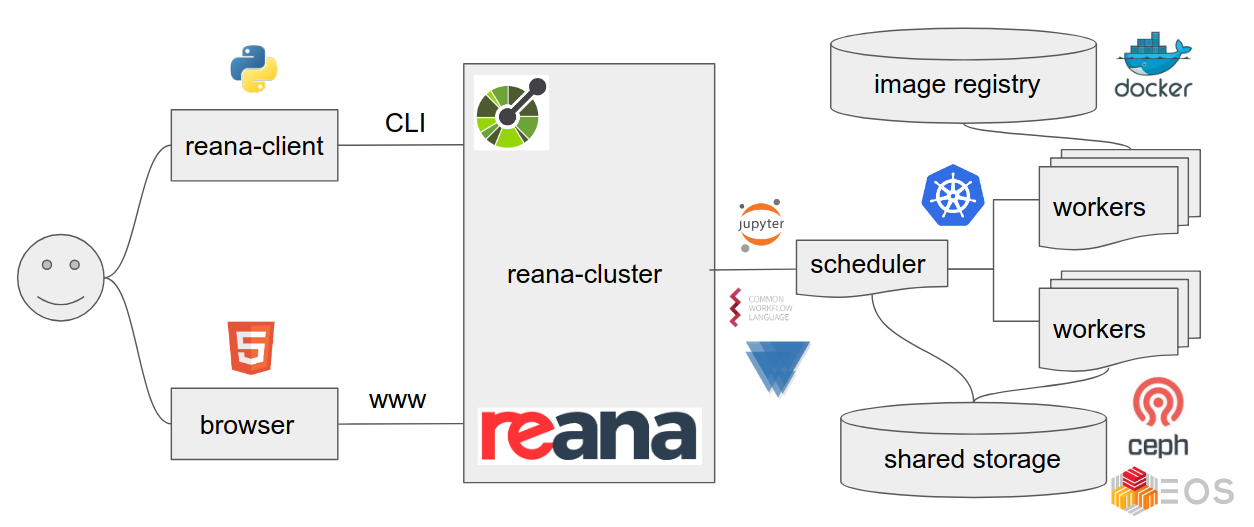
Analysis preservation ab initio
Preserving analysis code and processes after the publication is often too late. Key information and knowledge may be lost during the lengthy analysis process.
Making research reproducible from the start, in other words making research “preproducible”, makes analysis preservation easy.
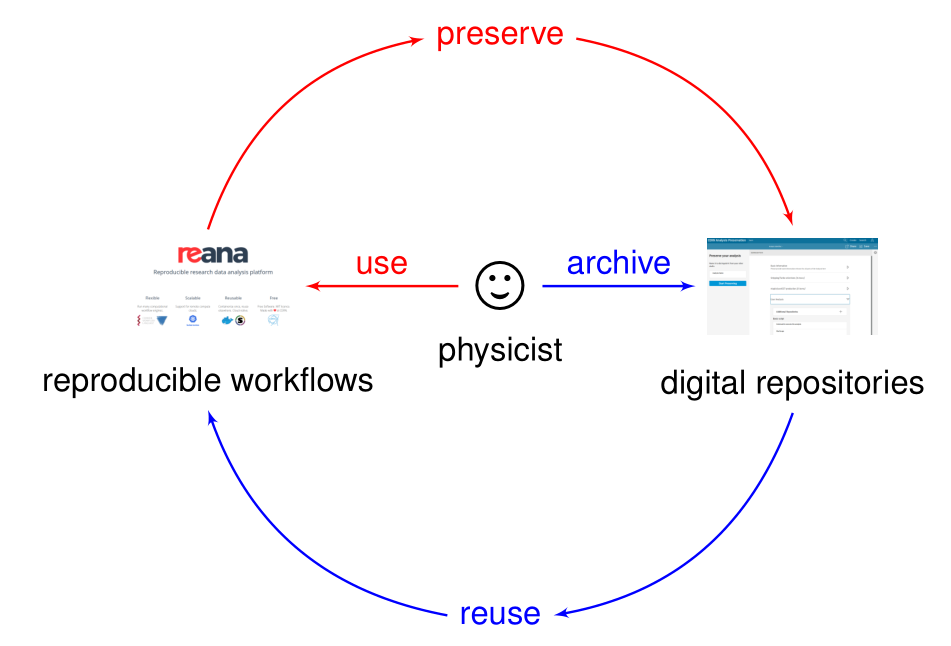
Preproducibility driving preservation: red-pill. Preservation driving reproducibility: blue pill.
Key Points
Workflow is the new data.
Data + Code + Environment + Workflow = Reproducible Analyses
Before reproducibility comes preproducibility