Developing parallel workflows
Overview
Teaching: 15 min
Exercises: 10 minQuestions
How to scale up and run thousands of jobs?
What is a DAG?
What is a Scatter-Gather paradigm?
How to run Yadage workflows on REANA?
Objectives
Learn about Directed Acyclic Graphs (DAG)
Understand Yadage workflow language
Practice running and inspecting parallel workflows
Overview
We now know how to develop reproducible analyses on small scale using serial workflows.
In this lesson we shall learn how to scale-up for real life work, which requires using paraller workflows.
Workflows as Directed Acyclic Graphs (DAG)
The computational analyses can be expressed as a set of steps where some steps depends on other steps before they can begin their computations. In other words, the computational steps as expressed as Directed Acyclic Graphs, for example:
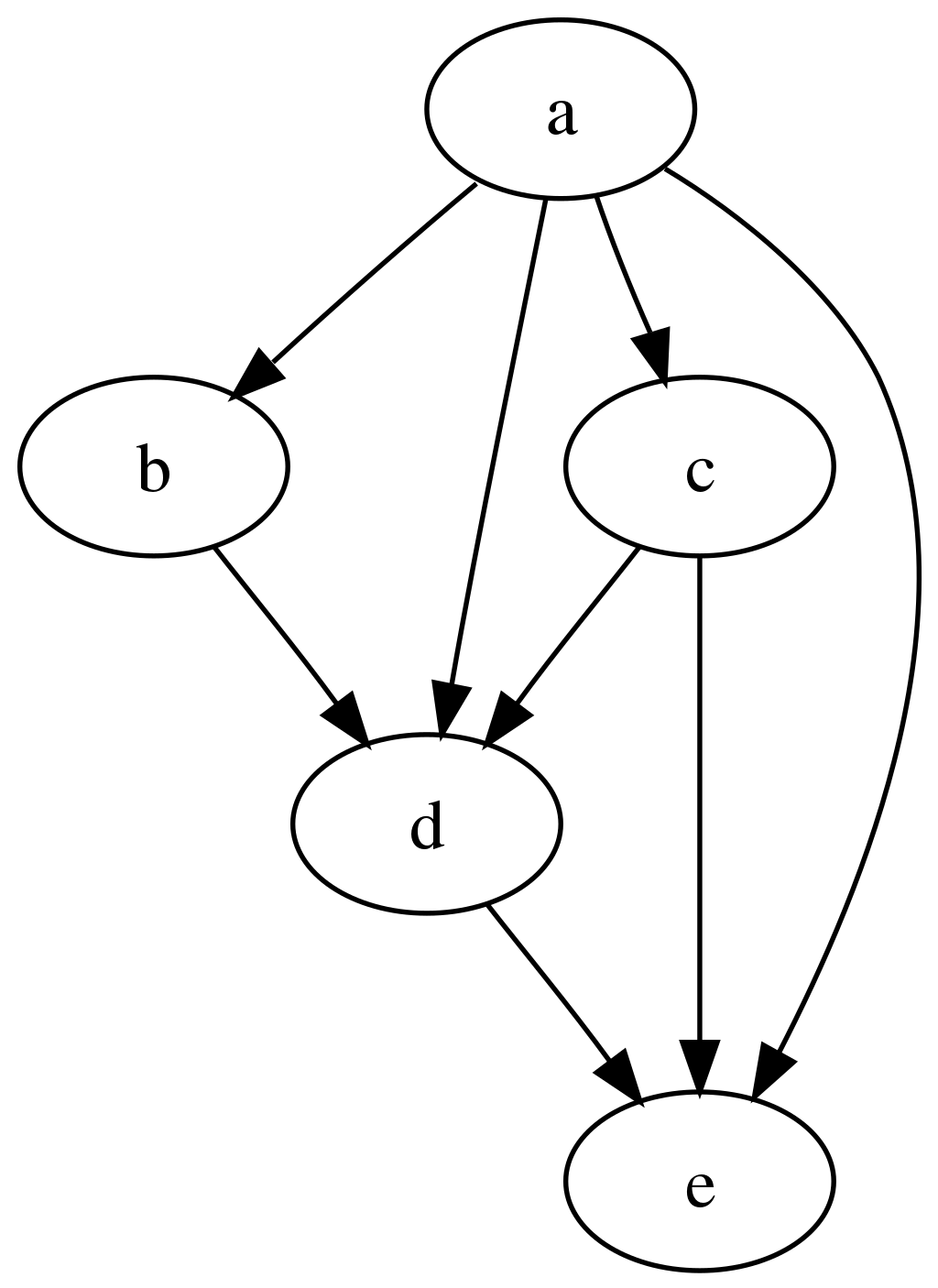
The REANA platform supports several DAG workflow specification languages:
- Common Workflow Language (CWL) originated in life sciences
- Yadage originated in particle physics
In this lesson we shall use the Yadage workflow specification language.
Yadage
Yadage enables to describe complex computational workflows. Let us start having a look at the Yadag e specification for the RooFit example we have used in the beginning episodes:
stages:
- name: gendata
dependencies: [init]
scheduler:
scheduler_type: 'singlestep-stage'
parameters:
events: {step: init, output: events}
gendata: {step: init, output: gendata}
outfilename: '{workdir}/data.root'
step:
process:
process_type: 'interpolated-script-cmd'
script: root -b -q '{gendata}({events},"{outfilename}")'
publisher:
publisher_type: 'frompar-pub'
outputmap:
data: outfilename
environment:
environment_type: 'docker-encapsulated'
image: 'reanahub/reana-env-root6'
imagetag: '6.18.04'
- name: fitdata
dependencies: [gendata]
scheduler:
scheduler_type: 'singlestep-stage'
parameters:
fitdata: {step: init, output: fitdata}
data: {step: gendata, output: data}
outfile: '{workdir}/plot.png'
step:
process:
process_type: 'interpolated-script-cmd'
script: root -b -q '{fitdata}("{data}","{outfile}")'
publisher:
publisher_type: 'frompar-pub'
outputmap:
plot: outfile
environment:
environment_type: 'docker-encapsulated'
image: 'reanahub/reana-env-root6'
imagetag: '6.18.04'
We can see that the workflow consists of two steps, gendata does not depending on anything
([init]) and fitdata depending on gendata. This is how linear workflows are expressed
in the Yadage language.
Running Yadage workflows
Let us write the above workflow as workflow.yaml in the RooFit example directory.
How can we run the example on REANA platform? We have to instruct REANA that we are going to use
Yadage as our workflow engine. We can do that by editing reana.yaml and specifying:
version: 0.6.0
inputs:
parameters:
events: 20000
gendata: code/gendata.C
fitdata: code/fitdata.C
workflow:
type: yadage
file: workflow.yaml
outputs:
files:
- fitdata/plot.png
We can run the example on REANA in the usual way:
$ reana-client run -w roofityadage -f reana-yadage.yaml
Exercise
Run RooFit example using Yadage workflow engine on the REANA cloud. Upload code, run workflow, inspect status, check logs, download final plot.
Solution
Nothing changes in the usual user interaction with the REANA platform:
$ reana-client create -w roofityadage -f ./reana-yadage.yaml $ reana-client upload ./code -w roofityadage $ reana-client start -w roofityadage $ reana-client status -w roofityadage $ reana-client logs -w roofityadage $ reana-client ls -w roofityadage $ reana-client download plot.png -w roofityadage
Physics code vs orchestration code
Note that it wasn’t necessary to change anything in our code: we simply modified the workflow definition and we could run the RooFit code “as is” using another workflow engine. This is a simple demonstration of the separation of concerns between “physics code” and “orchestration code”.
Parallelism via step dependencies
We have seen how serial workflows can be also expressed in Yadage syntax using step dependencies. Note that if dependency graph would have permitted, the workflow steps not depending on each other or on the results of previous computations could have been executed in parallel by the workflow engine – the physicist only has to supply knoweldge about which steps depend on which other steps and the workflow engine takes care about efficiently starting and scheduling tasks.
HiggsToTauTau analysis: simple version
Let us demonstrate how to write simple Yaage workflow for the HiggsToTauTau analysis using simple step dependencies.
The workflow looks like:
stages:
- name: skim
dependencies: [init]
scheduler:
scheduler_type: singlestep-stage
parameters:
input_dir: {step: init, output: input_dir}
output_dir: '{workdir}/output'
step: {$ref: 'steps.yaml#/skim'}
- name: histogram
dependencies: [skim]
scheduler:
scheduler_type: singlestep-stage
parameters:
input_dir: {step: skim, output: skimmed_dir}
output_dir: '{workdir}/output'
step: {$ref: 'steps.yaml#/histogram'}
- name: fit
dependencies: [histogram]
scheduler:
scheduler_type: singlestep-stage
parameters:
histogram_file: {step: histogram, output: histogram_file}
output_dir: '{workdir}/output'
step: {$ref: 'steps.yaml#/fit'}
- name: plot
dependencies: [histogram]
scheduler:
scheduler_type: singlestep-stage
parameters:
histogram_file: {step: histogram, output: histogram_file}
output_dir: '{workdir}/output'
step: {$ref: 'steps.yaml#/plot'}
where steps are expressed as:
skim:
process:
process_type: 'interpolated-script-cmd'
script: |
mkdir {output_dir}
bash skim.sh {input_dir} {output_dir}
environment:
environment_type: 'docker-encapsulated'
image: gitlab-registry.cern.ch/awesome-workshop/awesome-analysis-eventselection-stage3
imagetag: master
publisher:
publisher_type: interpolated-pub
publish:
skimmed_dir: '{output_dir}'
histogram:
process:
process_type: 'interpolated-script-cmd'
script: |
mkdir {output_dir}
bash histograms_with_custom_output_location.sh {input_dir} {output_dir}
environment:
environment_type: 'docker-encapsulated'
image: gitlab-registry.cern.ch/awesome-workshop/awesome-analysis-eventselection-stage3
imagetag: master
publisher:
publisher_type: interpolated-pub
publish:
histogram_file: '{output_dir}/histograms.root'
plot:
process:
process_type: 'interpolated-script-cmd'
script: |
mkdir {output_dir}
bash plot.sh {histogram_file} {output_dir}
environment:
environment_type: 'docker-encapsulated'
image: gitlab-registry.cern.ch/awesome-workshop/awesome-analysis-eventselection-stage3
imagetag: master
publisher:
publisher_type: interpolated-pub
publish:
datamc_plots: '{output_dir}'
fit:
process:
process_type: 'interpolated-script-cmd'
script: |
mkdir {output_dir}
bash fit.sh {histogram_file} {output_dir}
environment:
environment_type: 'docker-encapsulated'
image: gitlab-registry.cern.ch/awesome-workshop/awesome-analysis-statistics-stage3
imagetag: master
publisher:
publisher_type: interpolated-pub
publish:
fitting_plot: '{output_dir}/fit.png'
The workflow definition is similar to that of the Serial workflow, and, as we can see, it can already lead to certain parallelism, because the fitting step and the plotting step can run simultaneously once the histograms are produced.
The graphical representation of the workflow is:
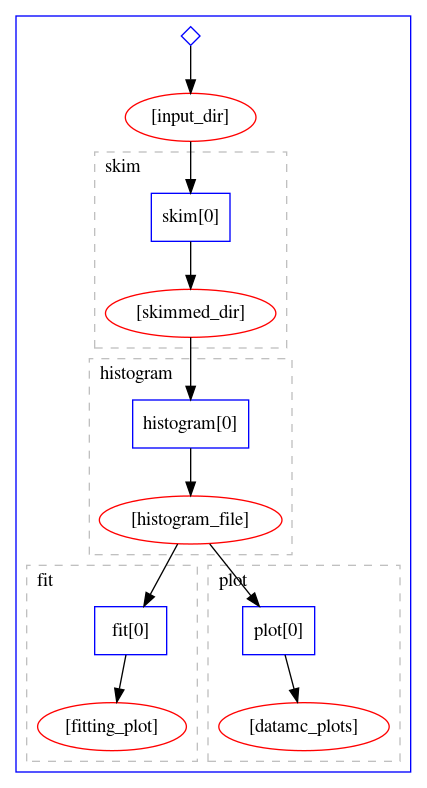
Let us try to run it on REANA cloud.
Exercise
Run HiggsToTauTau analysis example using simple Yadage workflow version. Take the workflow definition listed above, write corresponding
reana.yaml, and run the example on REANA cloud.
Solution
$ vim workflow.yaml # take contents above and store it as workflow.yaml $ vim steps.yaml # take contents above and store it as steps.yaml $ vim reana.yaml # this was the task $ cat reana.yaml version: 0.6.0 inputs: parameters: input_dir: root://eospublic.cern.ch//eos/root-eos/HiggsTauTauReduced workflow: type: yadage file: workflow.yaml outputs: files: - fit/output/fit.png
Parallelism via scatter-gather paradigm
A useful paradigm of workflow languages is a “scatter-gather” behaviour where we instruct the workflow engine to run a certain step over a certain input array in parallel as if each element of the input were a single item input (the “scatter” operation). The partial results processed in parallel are then assembled together (the “gather” operation). The “scatter-gather” paradigm allows to express “map-reduce” operations with a minimal of syntax without having to duplicate workflow code or statements.
Here is an example of scatter-gather paradim in the Yadage language:
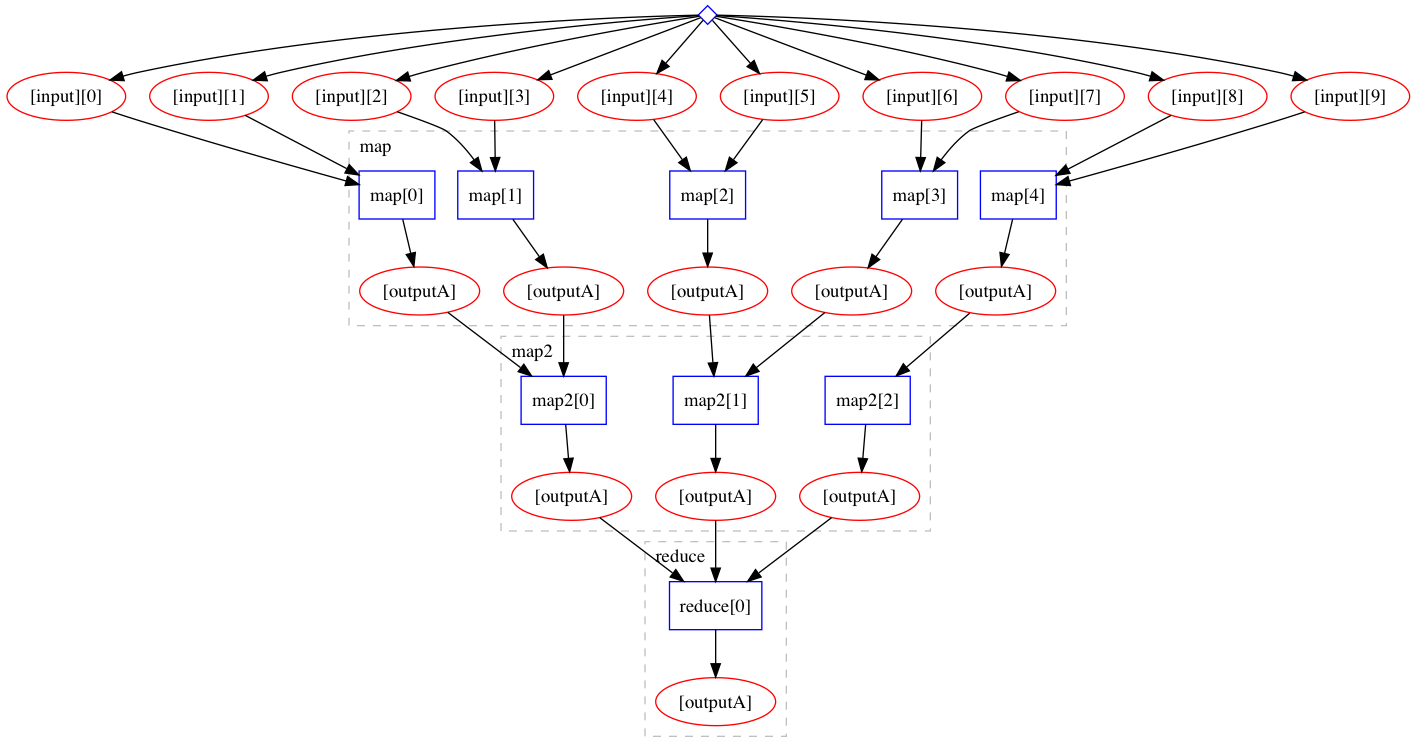
expressed as:
stages:
- name: map
dependencies: [init]
scheduler:
scheduler_type: multistep-stage
parameters:
input: {stages: init, output: input, unwrap: true}
batchsize: 3
scatter:
method: zip
parameters: [input]
- name: map2
dependencies: [map]
scheduler:
scheduler_type: multistep-stage
parameters:
input: {stages: map, output: outputA, unwrap: true}
batchsize: 2
scatter: ...
- name: reduce
dependencies: [map2]
scheduler:
scheduler_type: singlestep-stage
parameters:
input: {stages: 'map2', output: outputA}
Note the “scatter” happening over “input” with a wanted batch size.
In the next episode we shall see how the scatter paradigm can be used to speed up the HiggsToTauTau workflow using more parallelism.
Key Points
Computational analysis is a graph of inter-dependent steps
Fully declare inputs and outputs for each step
Use Scatter/Gather or Map/Reduce to avoid copy-paste coding